
My interests are at the interface of machine learning and digital health. Currently, I am a scientist at Apple. Previously I worked on applications of machine learning in healthcare at Philips Research with a special interest in bio-signal processing, patient similarity, and incorporating prior knowledge into AI models. I obtained my PhD in neural engineering and was an algorithms scientist at Stitch Fix where I worked on deep learning computer vision systems.
Research topics
1. Bio-signal processing
I specialize in deep learning algorithms for physiological signals collected from wearable devices, including electrocardiograms (ECG), photoplethysmography (PPG), phonocardiograms (PCG, heart sounds), and arterial blood pressure (ABP). Many of these solutions have been productionized and come in first place at the PhysioNet Challenges.See publications below.
2. Disease Phenotyping
I built realtime clinical risk algorithms that are deployed on patient monitors and central stations.
-
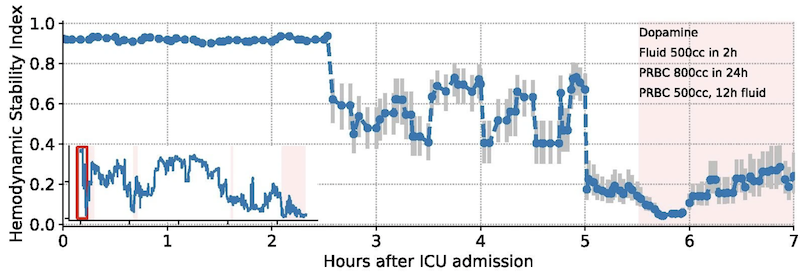
Early warning systems like the Hemodynamic Stability Index identifies ICU patients that will need fluids or pressors in the near future. The HSI model is designed for Philips bedside patient monitors and is the flagship machine learning model in hemodynamics analytics products. This work was extended with federated learning, a privacy preserving technique that was used to train the model across hundreds of ICUs.
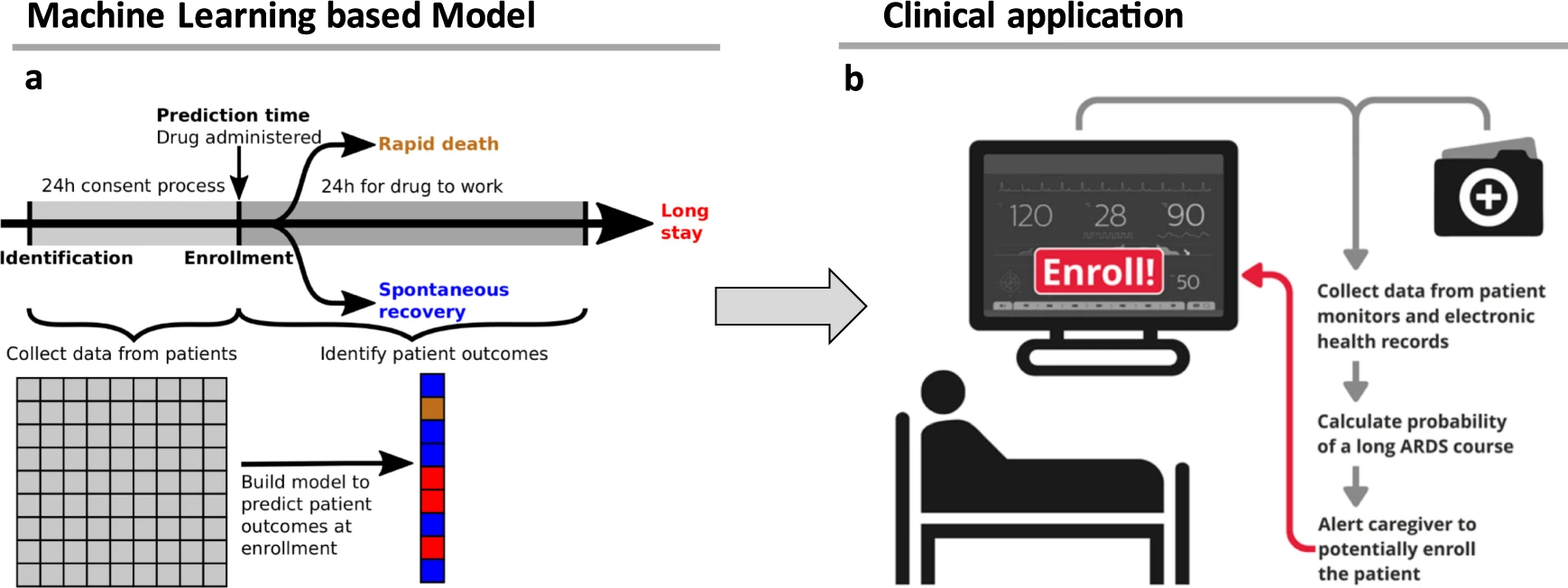
- Machine learning can be
used to improve clinical trial
design by finding the right patient at the right time. I led a collaboration with Bayer
Pharmaceuticals to optimize clinical trials investigating drugs to treat respiratory distress.
3. Patient similarity
Patient similarity aims to find cohorts based on vital signs, labs, medical history, and treatments for case-based reasoning.
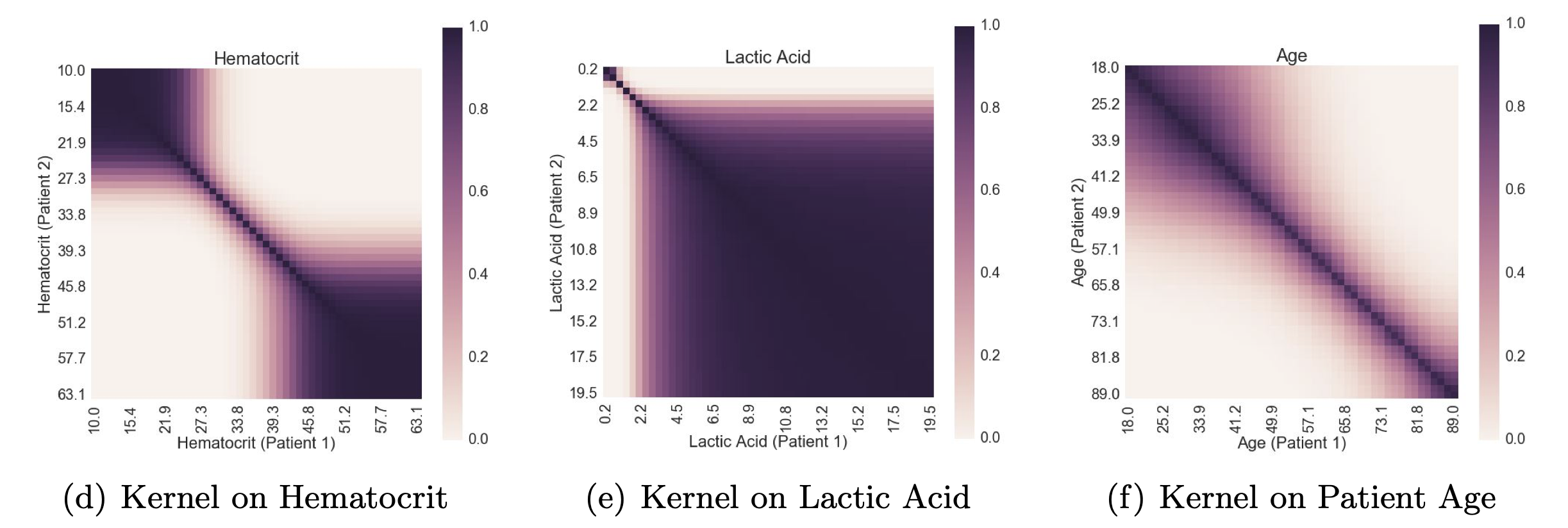
- How can we learn useful similarity functions that compare physiological state between patients? We
developed a
similarity metric to find
patients-like-me in
electronic health records.
-
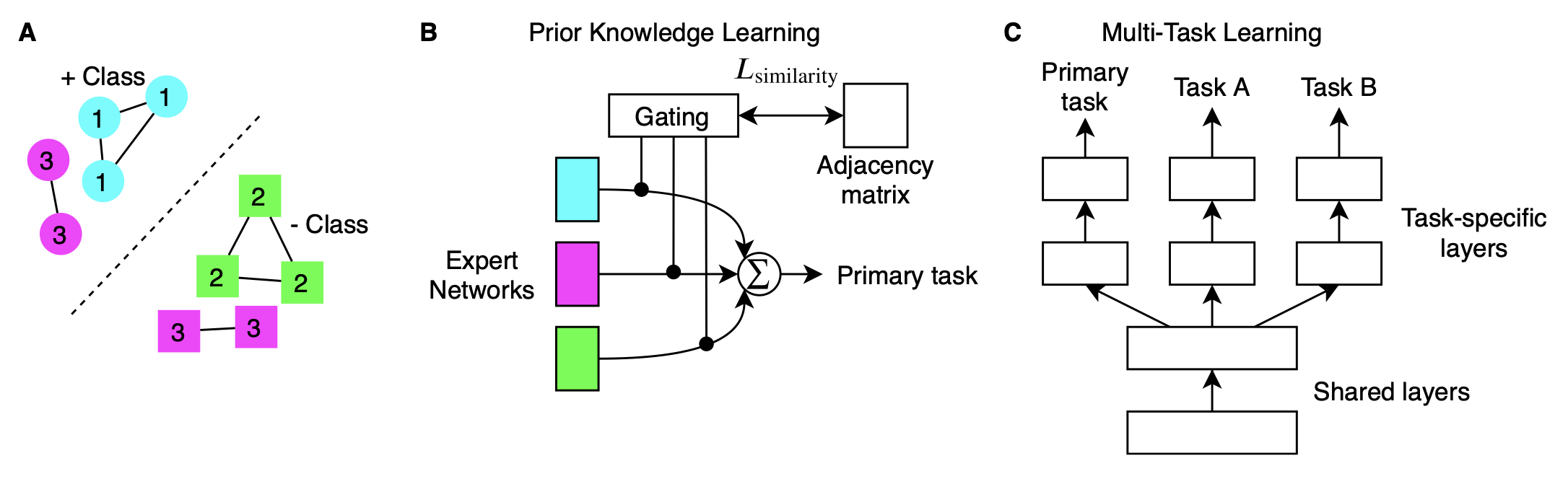
Prior medical knowledge is widely available in electronic health records but are not utilized because of practical constraints on data availability or cost of acquiring the data to make inferences. I developed a a novel approach to learn from metadata called prior knowledge-guided learning using a mixture-of-experts model where gating probabilities are tuned by a nearest neighbors graph adjacency matrix.
4. Interpretable machine learning
A key requirement of clinical models is that they have to be explainable so that the nurse or clinician can understand the risk factors. The models should also be editable so we can identify when then algorithm makes an error and fix it before deployment.
-
I created the Interpretable-RNN, which is an additive sequence learning model that achieves both high accuracy and is fully glassbox.
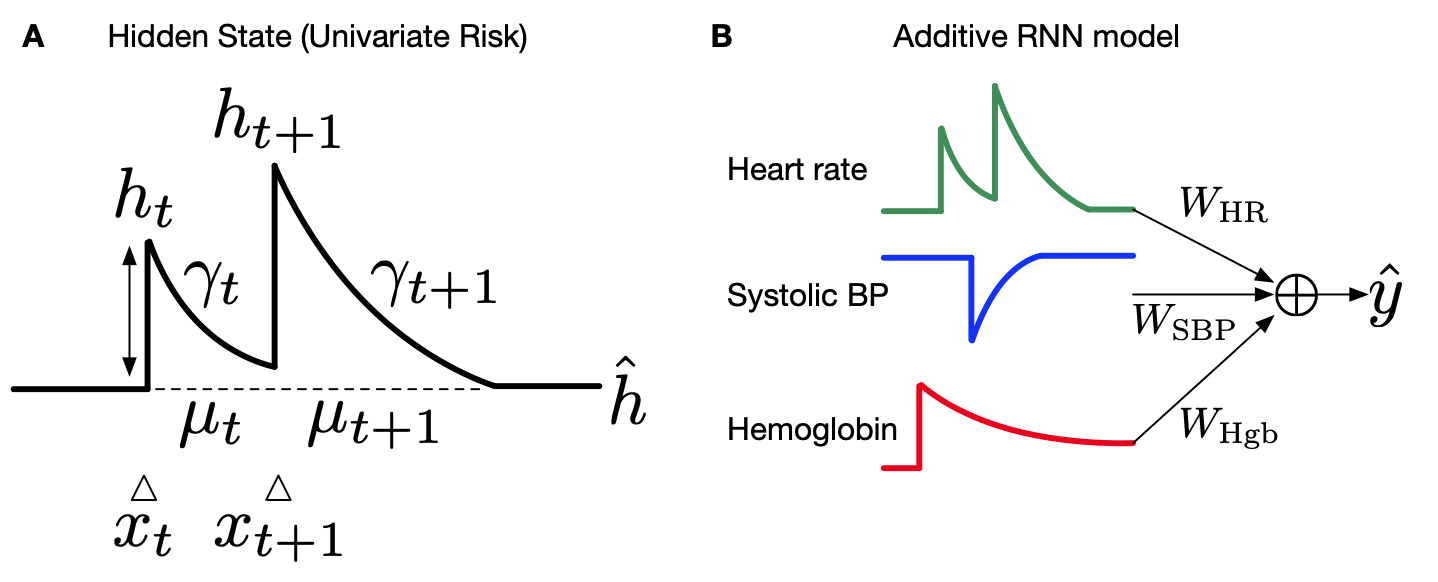
-
I developed the neural decision tree model which is a neural representation of highly interpretable gradient boosted decision trees. This allowed us to scale an interpretable class of models in a federated privacy preserving architecture.
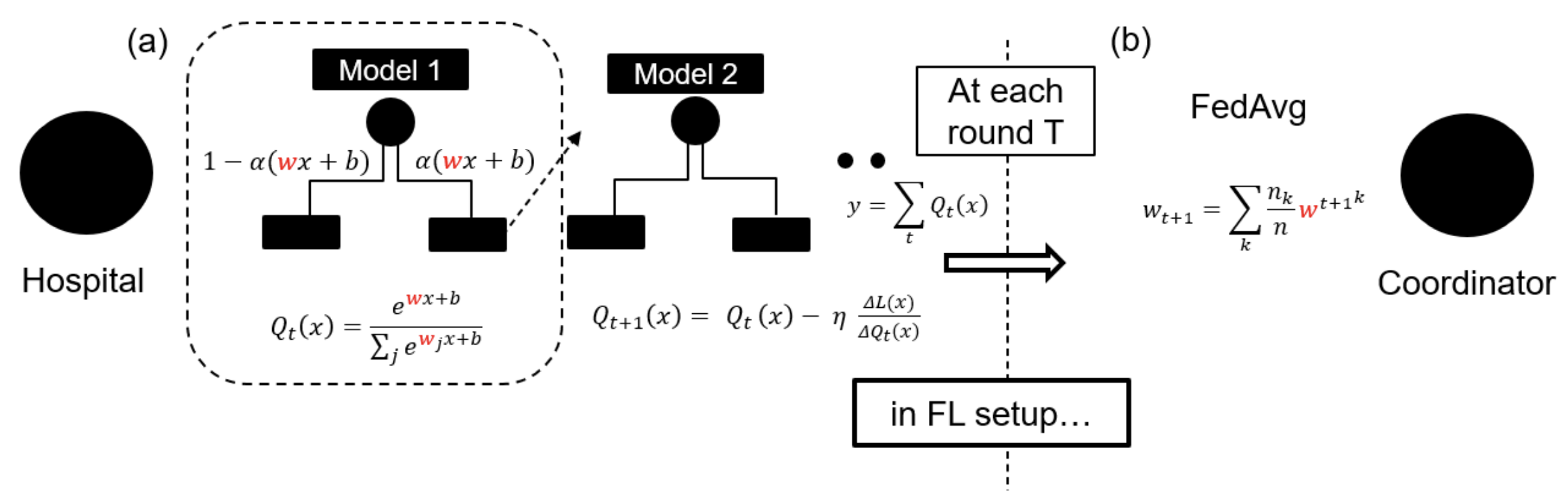
Publications
See all publications on Google Scholar